Problem Set 14: Gene Extraction
Web-CAT: Submit Java programs to this automated grading platform.
Task Outline
- Due Date: Monday, March 16, 2020
- Total Points: 10
- Implement a
Javaprogram that determines whether a gene is present in a DNA sequence.
Background Theory
- Deoxyribonucleic acid(DNA) is a complex biochemical macromolecule that carries genetic information for cellular life forms and some viruses. DNA is also the mechanism through which genetic information from parents is passed on during reproduction.
- DNA consists of long chains of chemical compounds called nucleotides. Four nucleotides are present in DNA: Adenine(A), Cytosine(C), Guanine(G), and Thymine(T). DNA has a double-helix structure(see diagram below) containing complementary chains of these four nucleotides connected by hydrogen bonds.
- Certain regions of the DNA are called genes. Most genes encode instructions for building proteins (these are called protein-coding genes). These proteins are responsible for carrying out most of the life processes of the organism.
- Nucleotides in a gene are organized into codons. Codons are groups of three nucleotides and are written as the first letters of their nucleotides(e.g., TAC or GGA). Each codon uniquely encodes a single amino acid, a building block of proteins.
- The process of building proteins from DNA has two major phases called transcription and translation, in which a gene is replicated into an intermediate form called mRNA, which is then processed by a structure called a ribosome to build the chain of amino acids encoded by the codons
of the gene.
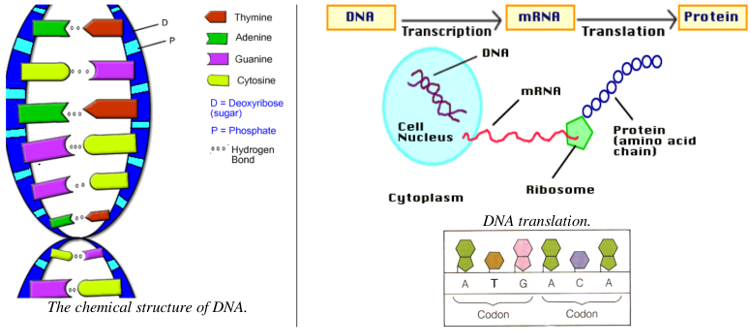
- The sequences of DNA that encode proteins occur between a start codon(which we will assume to be ATG) and a stop codon(which is any of TAA, TAG, or TGA). Not all regions of DNA are genes; large portions that do not lie between a valid start and stop codon are called intergenic DNA and have other(possibly unknown) functions.
- Computational biologists examine large DNA data files to find patterns and important information, such as which regions are genes. Sometimes, they are interested in the percentages of mass accounted for by each of the four nucleotide types. Often, high percentages of Cytosine(C) and Guanine(G) are indicators of important genetic data.
- For example, consider the following DNA sequence:
GTCATGCTACGTACGTATCGAGTCTCGTAACTG
- The start codon is
ATG, and the stop codon for this example isTAA. In order for this to qualify as a valid gene, it must have a minimum of 9 nucleotides between the start and stop codons. This particular example has 21 nucleotides between its start and stop codons, so it does qualify.
GTC[ATG]CTACGTACGTATCGAGTCTCG[TAA]CTG
- The nucleotide sequence that is present between the start and stop codons is as follows:
GTC[ATGCTACGTACGTATCGAGTCTCGTAA]CTG
- Now, we must calculate the percentage composition of Cytosine(C) and Guanine(G) in the nucleotide sequence, also known as the CG ratio.
\mbox{CG ratio} = 1.0\times\frac{\mbox{quantity of C and G nucleotides}}{\mbox{quantity of all nucleotides}}
- In this case, the quantity of C and G nucleotides is 12. The count of all the nucleotides in the sequence is 27. Therefore, the CG ratio is calculated as follows:
\mbox{CG ratio} = 1.0\times\frac{12}{27} \\
\mbox{CG ratio} = 0.444444
- Since the CG ratio is greater than 0.3, we can conclude that this nucleotide sequence is a valid, protein-coding gene.
- Note that we need to multiply this ratio by 1.0, in order to force a decimal division.
Hints
- Your program must examine each of the provided DNA sequences, and determine if a valid, protein-coding gene is present. This gene must satisfy the following constraints:
- It must begin with a valid start codon: ATG
- It must end with a valid stop codon: TAA, TAG, or TGA
- It must contain at least 5 codons, including its initial start codon, and its final stop codon.
- The size of the gene must be a multiple of 3.
- The nucleotides Cytosine(C) and Guanine(G) must make up at least 30% of the gene's total composition.
- First, you should examine the provided DNA sequence for the index of the start codon, ATG. If this start codon is not present in the DNA sequence, then you simply cannot have any gene in there.
- Note that all of the DNA sequences under consideration will only contain zero or one ATG start codons. In other words, we will not consider cases where the DNA sequence contains more than one gene.
- Once you have the index of the ATG start codon, you should scan through the rest of the DNA sequence for a stop codon. There are only two possibilities here: either you find a stop codon, or you reach the end of the DNA sequence(in which case you didn't find a stop codon).
- As you are scanning through the DNA sequence, you should probably examine it in string chunks of size three. This makes it easy to compare each string chunk with each of the stop codon varieties.
- Once you have found a stop codon, you should check that the gene contains at least 5 codons in total, including the start and stop codons.
- Then, you should perform a simple calculation to verify that at least 30% of the gene's nucleotides are Cytosine(C) or Guanine(G).
- At various points in your program, especially during the evaluation of certain conditional statements, you may come to the conclusion that you don't have a valid gene. In such cases, there is no need to continue with the rest of the program, and you can simply exit the method with the statement:
return "INVALID"
Code Distribution
| Description | File Size | File Name |
|---|---|---|
Java Source Code for Gene Extraction |
6.6KB | pset14.zip |
Contents of pset14.zip:
PSet14GeneExtraction/
├── GeneExtraction.class
├── GeneExtraction.ctxt
├── GeneExtraction.java
├── GeneExtractionJUnitTest.class
├── GeneExtractionJUnitTest.ctxt
├── GeneExtractionJUnitTest.java
├── package.bluej
└── README.TXT
Specification
- Write a
Javaprogram in the fileGeneExtraction.javathat determines whether a valid gene is present in a DNA sequence. If this is the case, then your program should return the sequence of nucleotide letters that represents the gene. Otherwise, it should return the string:INVALID - You will write your solution in a function called
geneSequence(String dna)right below the place where it says:YOUR CODE HERE - When the function call
geneSequence("GTCATGCTACGTACGTATCGAGTCTCGTAACTG")is executed, the output of the program should be:ATGCTACGTACGTATCGAGTCTCGTAA
Testing
- Click on the Run Tests button to execute the
JUnittest bench.JUnitindicates a successful test with a green bar, and an unsuccessful test with a red bar.
Submission
- Upload the file
GeneExtraction.javato the Web-CAT automated grading platform.